Tools
We develop and maintain freely available, open source software implementations of genomic data science tools.
Featured

Deconvolution of cell types from MeDIP-seq

Regional Association of Methylome variability with the Exposome and geNome

R package for detecting variably methylated regions (VMRs) from single-cell methylation sequencing

R package to Perform Bayesian Decision Curve Analysis for clinical prediction models and diagnostic tests

an R/Bioconductor package for differential methylation analysis using bulk whole genome bisulfite sequencing data

an R/Bioconductor package for differential distribution analysis using single-cell RNA-sequencing data
More
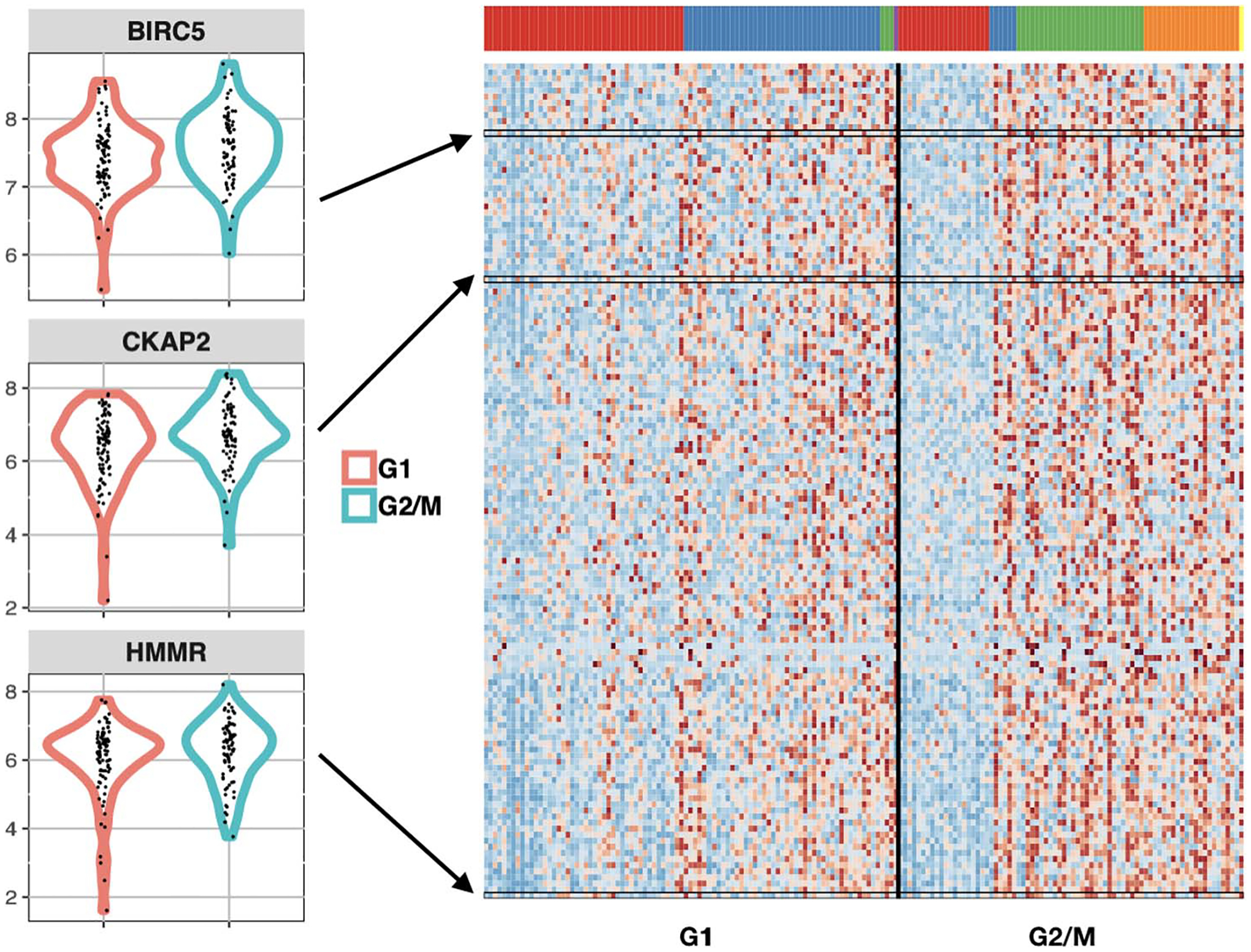
an R package to analyze changes in the distribution of single-cell expression data between two experimental conditions with multiple cell types

R/Shiny application for exploring results from “A practical guide to methods controlling false discoveries in computational biology”

R/Bioconductor Experiment Package containing benchmark results from “A practical guide to methods controlling false discoveries in computational biology”

an R/Bioconductor package defining S4 Classes for storing Single Cell Data
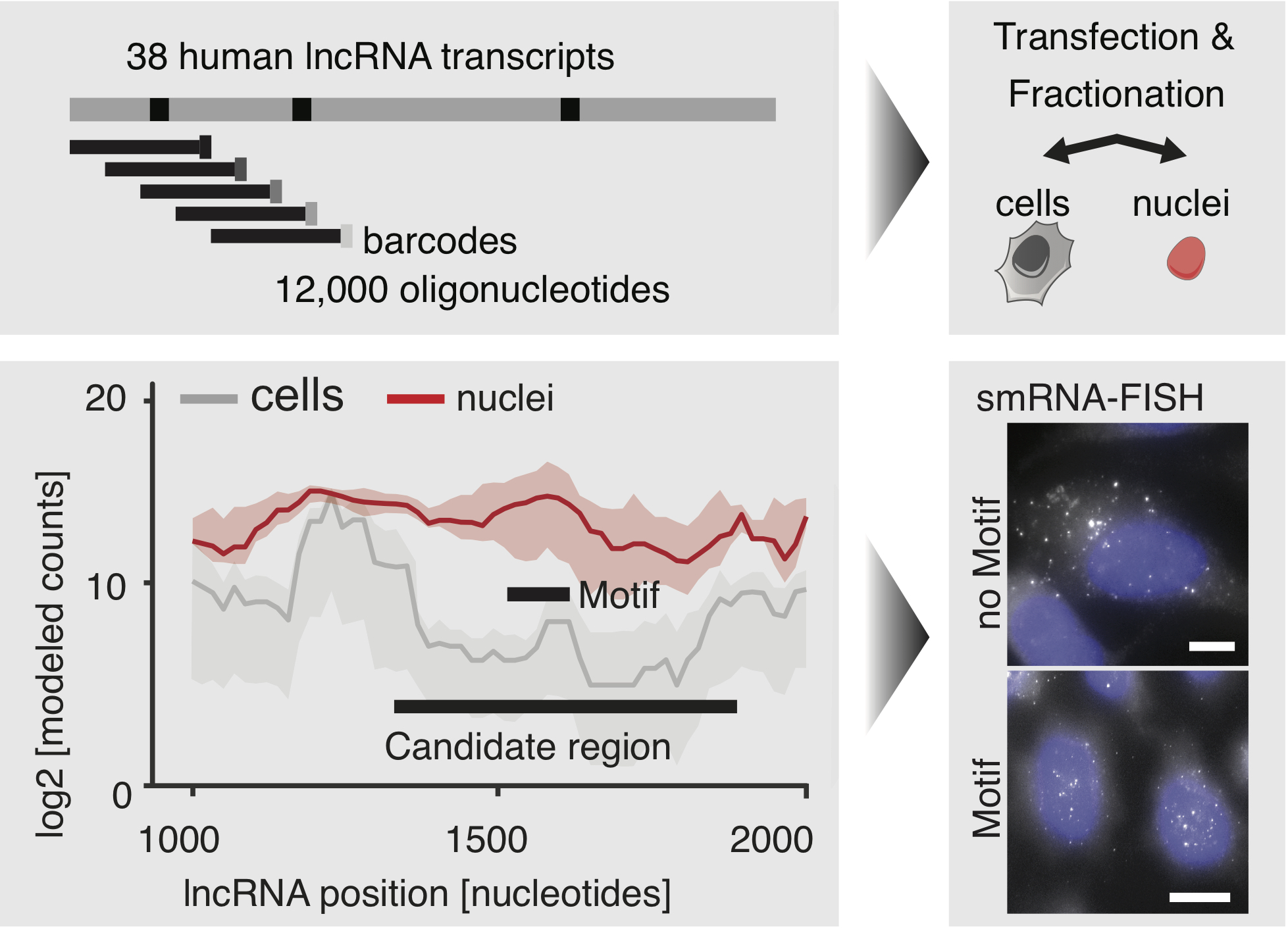
an R package for the analysis of tiled massively parallel reporter assays (MPRAs)
