Research
Our interdisciplinary research lies at the interface of statistics and genomics. With our methods development work, we aim to advance statistical methodology and computational tools for the analysis of high-dimensional biological data. Our work is also highly collaborative; we routinely work alongside experimentalists and clinicians to apply cutting edge bioinformatics tools to uncover new molecular signals in cancer, child health, and development. We are especially interested in advancing understanding of epigenetics - the study of how gene activity is controlled through mechanisms that don’t involve changes in the DNA sequence itself - at the genome scale. You can check out our publications listed below, or find them listed on Google Scholar.
2025


2024






2023

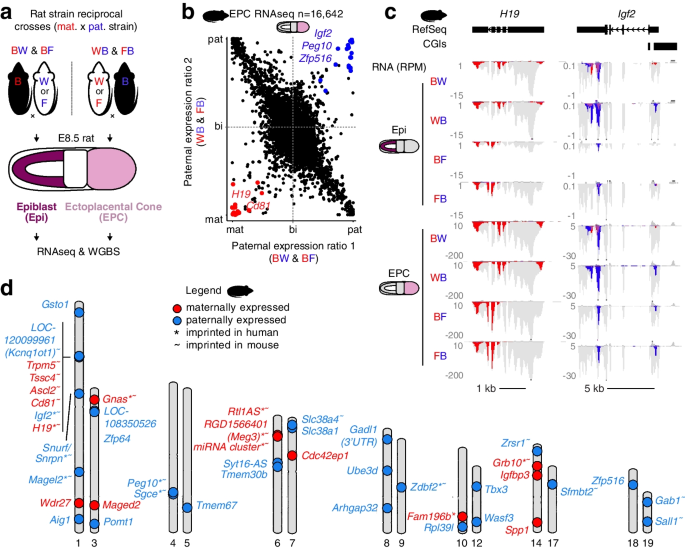
2022


2021
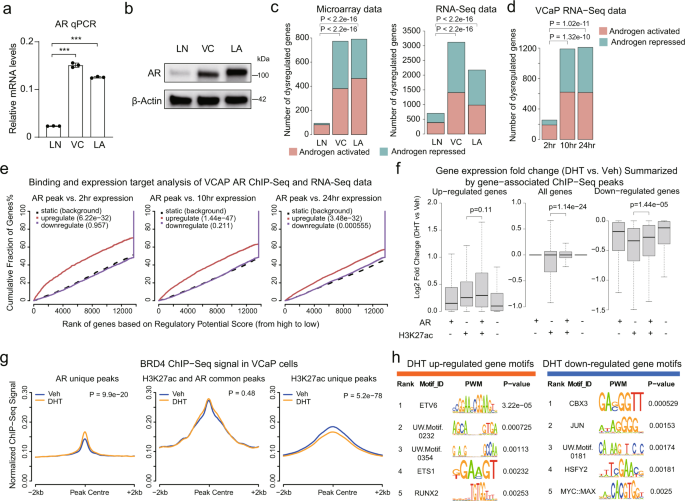
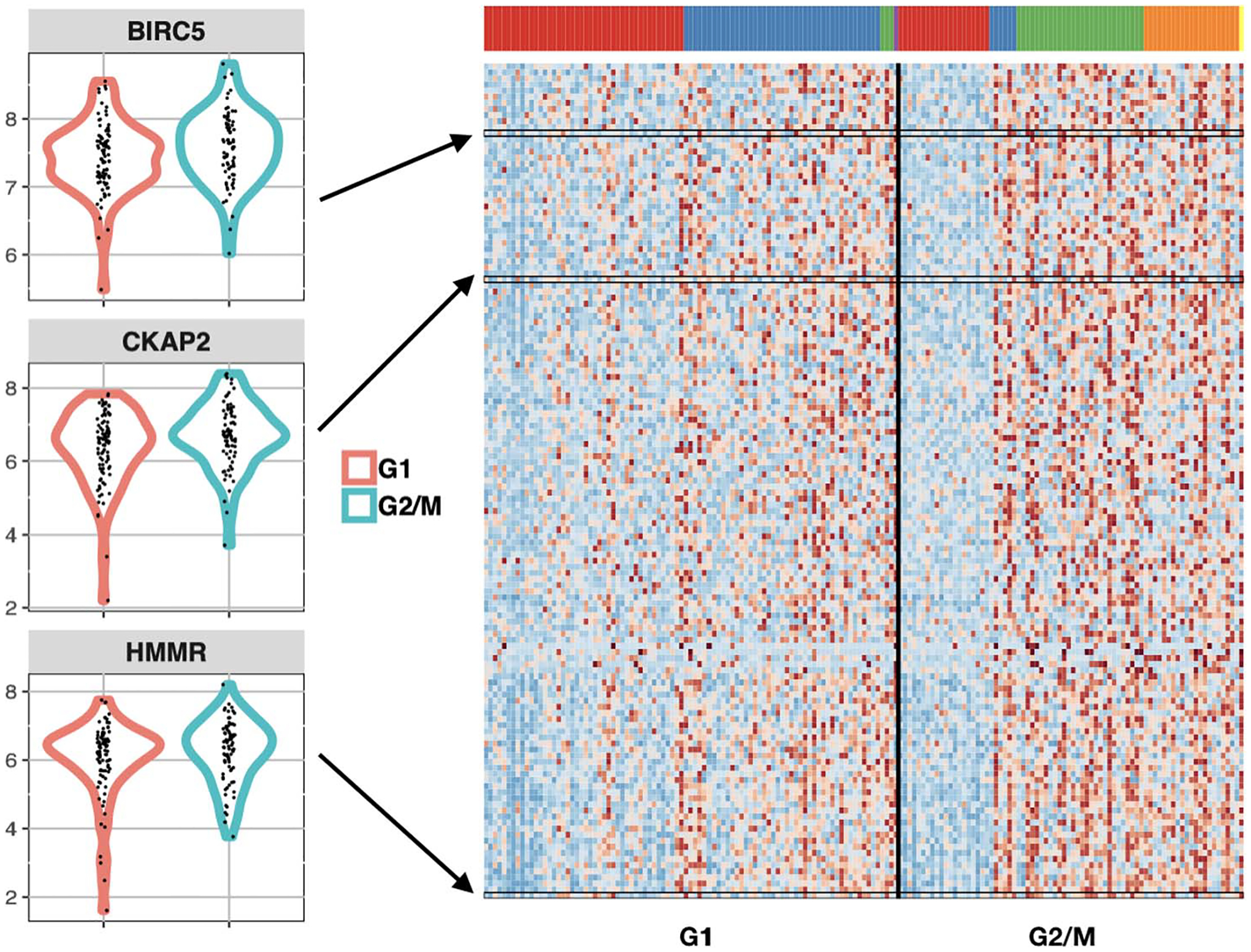

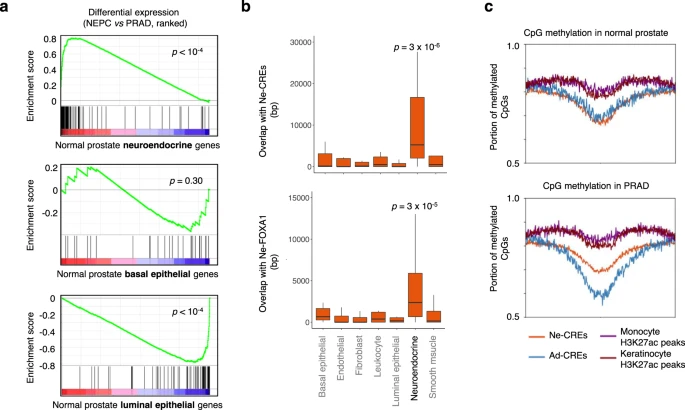
2020
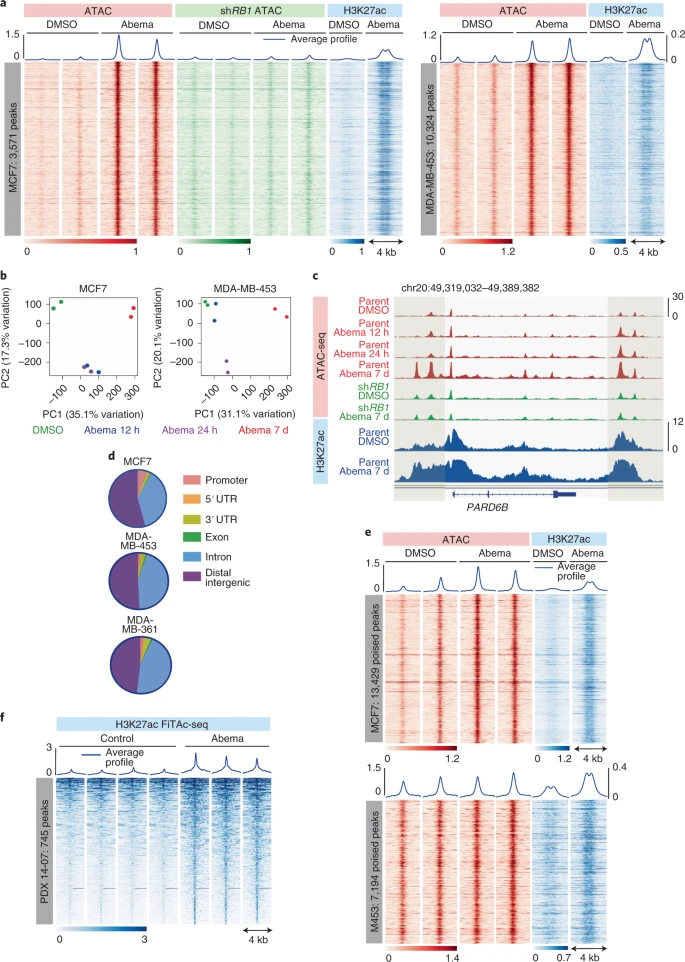


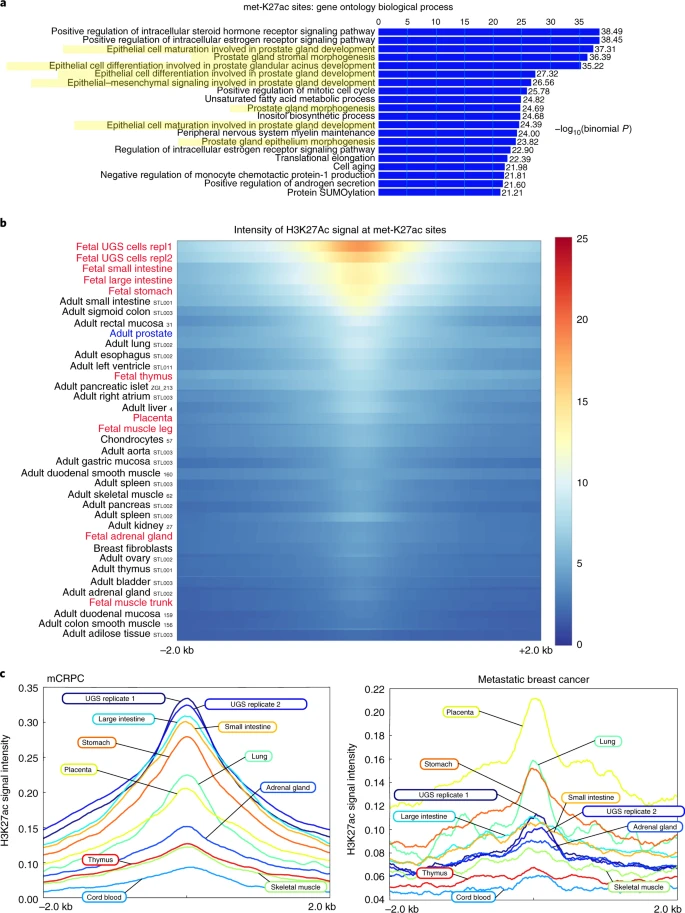
2019
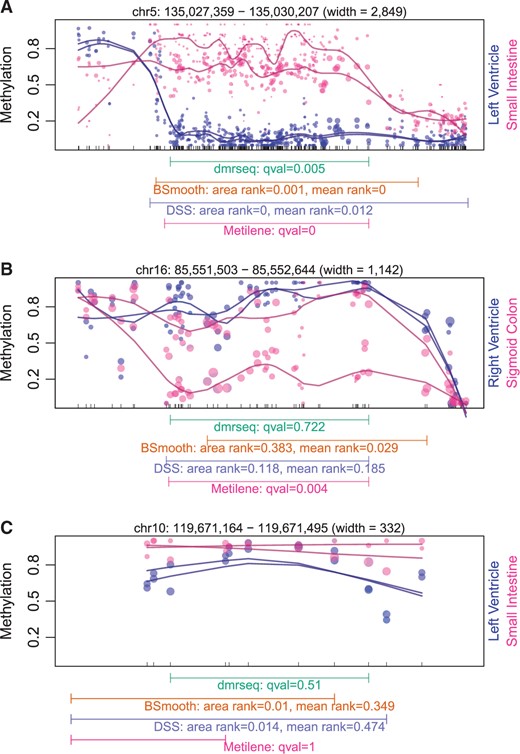

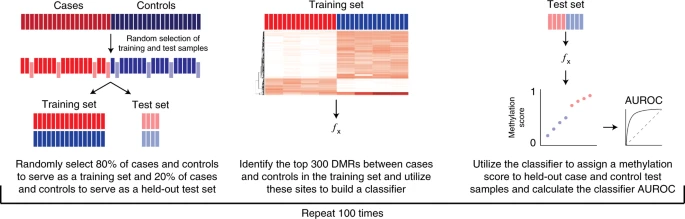
2018


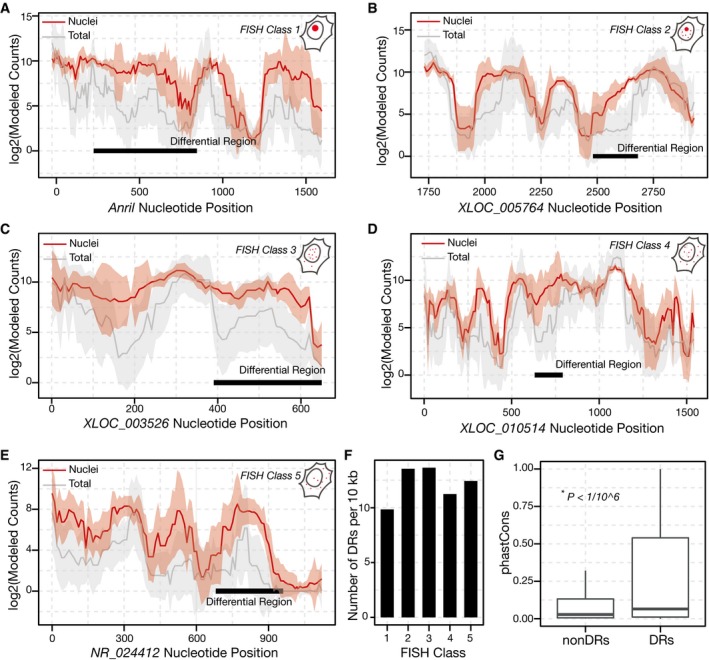
2017
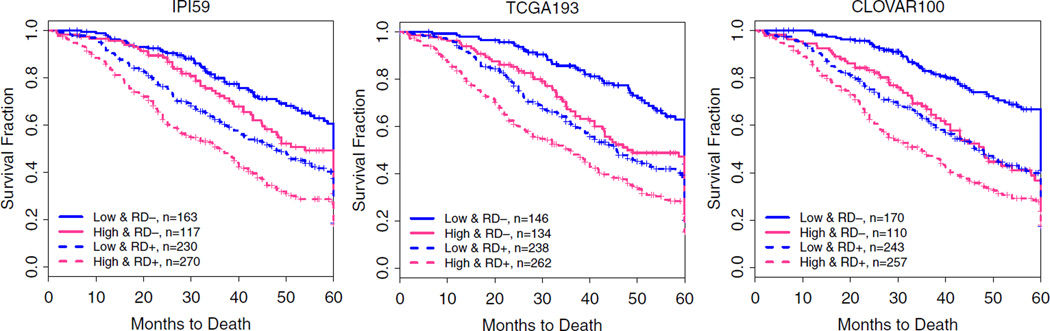
2016
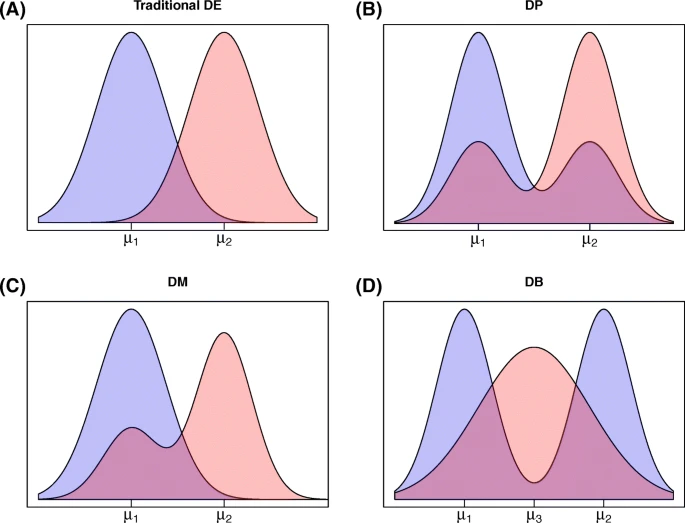
2015
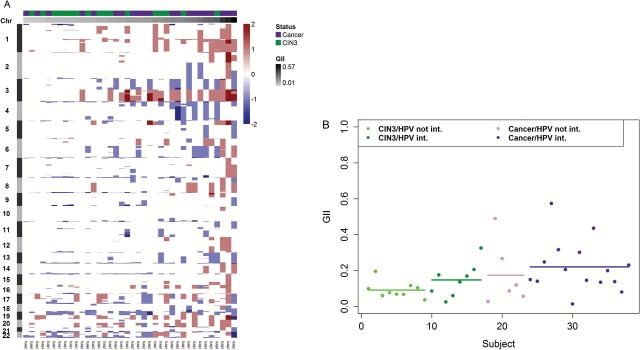

2014

2013

2012

2008

